| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: About this document ... Up: XPLOR Interface Manual Previous: Abbreviations
Index
- statement
- Syntax
- ( )
- Notation
- 1-4 interaction
- Syntax | Syntax | Syntax | Syntax | Files “parhcsdx.pro" and “tophcsdx.pro" | Intramolecular Interactions
- :==
- Notation


- Notation
 * *
* *
- Notation
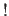
- Words
 * *
* *
- Wildcards
- @
- Control Statements
- @@
- Control Statements
- {}
- Words
- |
- Notation
- accessible surface area
- Accessible Surface Area
- angle database restraint
- Angle Database Restraint
- ANGLedb-statement
- Syntax
- ANIS statement
- Syntax - ANIS
- anomalous scattering
- Anomalous Scattering
- antidistance restraints
- Antidistance Restraints
- application
- Control Statements | Application Statements
- atom properties
- Syntax
- atom selection
- Atom Selection
- atom selection, fragile
- Atom Selection
- atom tag
- Syntax | Conformation vs. Residue Number
- atom-based parameters
- Parameter Statement | Examples of Atom-based Modifications
- atomic form factors
- Syntax of the Xrefin
- B-factor refinement
- Overall B-Factor Refinement | Individual B-Factor Refinement
- Backus-Naur notation
- Notation
- bond angle deviations
- Example: Print Bond Length
- bond length deviations
- Example: Print Bond Length
- bound smoothing
- Bound Smoothing
- boundary forces, deformable
- Deformable Boundary Forces
- Brookhaven Protein Data Bank
- Syntax
- building,hydrogens
- Building Hydrogen Positions
- carbohydrates,topologies and parameters
- Files “toph3.cho" and “param3.cho"
- carbon chemical shift
- Carbon Chemical Shift Restraints
- CARBon shift statement
- Syntax
- centroid search
- Intramolecular Interactions
- CHARMM trajectory format
- Binary Trajectory Files
- chemical shift anisotropy restraint
- Chemical Shift Anisotropy Restraint
- chemical shift, carbon
- Carbon Chemical Shift Restraints
- chemical shift, proton
- Proton Chemical Shift Restraints
- chiral centers
- Test for the Correct
- chirality
- Conformational Energy Terms | NMR Structure Determination
- chromophores,topologies and parameters
- Files “toph19.chromo" and “param19.chromo"
- COLLapse-statement
- Syntax
- completeness of diffraction data
- Syntax of the Xrefin
- conformational analysis
- Conformation vs. Residue Number
- conjugate gradient minimization
- Conjugate Gradient Minimization
- constraints interaction statement
- Energy Calculation between Selected
- constraints,coordinate
- Coordinate Constraints
- constraints,fixing atomic positions
- Fixing Atomic Positions
- constraints,fixing distances
- Fixing Distances
- constraints,SHAKE
- Fixing Distances
- control statement
- Control Statements
- coordinates
- Cartesian Coordinates
- coordinates, fractionalization
- Examples | Orthogonalization Convention
- coordinates, orthogonalization
- Examples | Orthogonalization Convention
- coordinates, unknown
- What to Do about
- coordinates,best fit
- Syntax
- coordinates,comparison set
- Syntax
- coordinates,manipulating
- Coordinate Statement
- coordinates,rms difference
- Syntax
- coordinates,rms difference (rmsd) analysis
- Rms Differences between Coordinates
- coordinates,rms difference analysis
- Average Structure and Rmsds
- coordinates,rms difference,pairwise
- Pairwise Rmsds
- coordinates,rotation and translation
- Syntax
- coordinates,writing
- Write Coordinate Statement
- COUPlings statement
- Syntax
- crystal packing
- Deviations from Ideality and
- crystallographic diffraction data
- Syntax of the Xrefin
- crystallographic refinement
- Crystallographic Refinement
- crystallographic refinement, atomic positions
- Positional Refinement
- crystallographic refinement, simulated annealing
- Crystallographic Refinement by Simulated
- crystallographic refinement, B-factors
- Overall B-Factor Refinement | Individual B-Factor Refinement
- crystallographic refinement,alternate conformations,disorder
- Alternate Conformations
- crystallographic refinement,bad contacts
- Check of Data and
- crystallographic refinement,initial structure check
- Check of Data and
- crystallographic refinement,linear correlation coefficient
- Crystallographic Target Functions
- crystallographic refinement,occupancies
- Grouped B-Factor and Occupancy
- crystallographic refinement,overview
- Crystallographic Refinement
- crystallographic refinement,phase accuracy
- Cross-validation: The Free
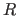
- crystallographic refinement,slow-cooling
- Crystallographic Refinement by Simulated
- crystallographic refinement,special positions
- Special Positions
- crystallographic refinement,target functions
- Crystallographic Target Functions
- crystallographic refinement,topologies and parameters
- Files “parallhdg.pro" and “topallhdg.pro"
- crystallographic refinement,weights
- Check of Data and
- CSA restraint
- Chemical Shift Anisotropy Restraint
- DANI-statement
- Syntax
- database restraints, dihedral
- Dihedral Angle Database Restraints
- database restraints, positional
- Residue-Residue Position/Orientation Database Restraint
- DCSA-statement
- Syntax
- deformable boundary forces
- Deformable Boundary Forces
- deletion of atoms
- Deleting Atoms
- deviation from ideality
- Deviations from Ideality and
- diffraction data
- Crystallographic Diffraction Data
- diffraction data, completeness
- Syntax of the Xrefin
- diffraction data, expanding
- Example: Expand a Data
- diffraction data, reducing
- Example: Expand a Data
- diffraction data,manipulation
- Manipulating Reflection Data
- diffraction data,scaling
- Crystallographic Target Functions | Syntax
- diffusion anisotropy restraints
- Diffusion Anisotropy Restraints
- dihedral angle restraints
- Dihedral Angle Restraints
- dihedral restraints, database
- Dihedral Angle Database Restraints
- dipolar couplings
- Residual Dipolar Couplings
- DIPOlar statement
- Syntax - XDIPolar or
- directory environment variables
- Commonly used UNIX variables
- display
- Control Statements
- distance geometry,van der Waals interactions
- Input Distances
- distance analysis
- Analysis of the Nonbonded
- distance difference matrix
- Distance Matrix Analysis
- distance geometry
- Distance Geometry | Implementation of Distance Geometry
- distance geometry, accuracy parameter
- Input Distances
- distance geometry,bond angle constraints
- Input Distances
- distance geometry,bound smoothing
- Bound Smoothing
- distance geometry,dihedral angle constraints
- Input Distances
- distance geometry,distance bounds
- Input Distances
- distance geometry,embedded substructures
- Distance Geometry
- distance geometry,embedding
- Embedding
- distance geometry,full-structure embedding
- Full-Structure Distance Geometry
- distance geometry,improper angle constraints
- Input Distances
- distance geometry,metrization
- Metrization
- distance geometry,pseudoatoms
- Pseudoatoms
- distance geometry,regularization of coordinates
- Regularization
- distance geometry,rigid group
- Syntax
- distance geometry,SA-regularization of structures
- SA-Regularization of DG-Structures
- distance geometry,scaling
- Scaling
- distance geometry,triangle inequalities
- Bound Smoothing
- distance matrix
- Distance Matrix Analysis
- distance restraints
- Distance Restraints
- distance restraints, restraining functions
- Choice of Restraining Functions
- distance restraints,ambiguous
- Expressing ambiguous restraints using
- distance restraints,averaging methods
- Choice of Averaging
- distance restraints,disulfide bonds
- Incorporation of Other Distance
- distance restraints,hydrogen bonds
- Incorporation of Other Distance
- distance restraints,reading
- Setup of Distance Restraints
- distance,between atoms
- Example: Pick Distance between
- disulfide bridge
- Example: Incorporation of Disulfide
- disulfide bridges
- NMR Structure Determination
- duplicating the molecular structure
- Duplicating the Molecular Structure
- electron density map
- Electron Density Maps
- electron density map,
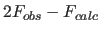 map
map
- Example: Computation of a
- electron density map,map file
- Electron Density Map File
- electron density map,omit map
- Example: Computation of an
- electron density map,SA-refined omit map
- Example: Computation of an
- embedding
- Embedding
- empirical energy function
- Empirical Energy Functions
- energy analysis, conformational terms
- Geometric and Energetic Analysis
- energy analysis,nonbonded terms
- Analysis of the Nonbonded
- energy analysis,setting the weight
- Syntax
- energy evaluation
- Energy Statement
- energy function
- Energy Function | Empirical Energy Functions
- energy minimization
- Energy Minimization
- energy, bond
- Conformational Energy Terms
- energy, bond angle
- Conformational Energy Terms
- energy, conformational
- Conformational Energy Terms
- energy, dihedral angle
- Conformational Energy Terms
- energy, electrostatic
- Electrostatic Function
- energy, hydrogen bond
- The Explicit Hydrogen-Bond Term
- energy, improper angle
- Conformational Energy Terms
- energy, nonbonded
- Nonbonded Energy Terms
- energy, torsion angle
- Conformational Energy Terms
- energy, van der Waals
- Van der Waals Function
- energy,analysis
- Energy Calculation between Selected
- energy,crystallographic symmetry
- Crystallographic Symmetry Interactions
- energy,interaction between selected atoms
- Energy Calculation between Selected
- energy,intermolecular
- Crystallographic Symmetry Interactions | Non-crystallographic Symmetry Interactions
- energy,intramolecular
- Intramolecular Interactions
- energy,non-crystallographic symmetry
- Non-crystallographic Symmetry Interactions
- energy,pick
- Syntax of the Pick
- energy,print
- Syntax of the Print
- energy,second function
- Energy Calculation between Selected | Syntax
- energy,turning on or off
- Turning Energy Terms On
- equal sign
- Notation
- Eulerian angles
- 3
 3 Matrices
3 Matrices
- evaluate
- Evaluate Statement
- example files,on-line
- Electronic Location of Example
- fast Fourier transformation
- Crystallographic Target Functions | Syntax of the Xrefin
- figure of merit
- Crystallographic Target Functions | Syntax of the Xrefin | Syntax
- filenames
- Filenames
- flags
- Turning Energy Terms On
- for ...in
- Control Statements
- force-field
- Topology and Parameter Files
- fractional coordinates
- Orthogonalization Convention
- fractionalized coordinates
- Examples
- fragile atom selection
- Atom Selection
- free
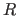 value
value
- Cross-validation: The Free
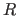
- Friedel mates
- Syntax of the Xrefin
- geometric analysis
- Geometric and Energetic Analysis | Deviations from Ideality and
- geometry,pick
- Syntax of the Pick
- geometry,print
- Syntax of the Print
- harmonic restraints
- Harmonic Coordinate Restraints
- HBDA-statement
- Syntax
- HBDB-statement
- Syntax
- heavy-side function
- Syntax
- help on-line
- On-line HELP and Query
- histidine protonation
- Example: Modification of the
- hydrogen bond database restraint
- Hydrogen Bond Database Restraint
- hydrogen bond geometry restraint
- Hydrogen Bond Geometry Restraint
- hydrogen building
- Building Hydrogen Positions
- hydrogen-bond donors and acceptors
- The Explicit Hydrogen-Bond Term
- hydrogen-bonding parameters
- Syntax
- hydrogens
- Syntax | Building Hydrogen Positions
- hydrogens,naming convention
- Convention for Hydrogen Names
- if ...then
- Control Statements
- inertia tensor
- Rigid-Body Coordinate Space
- interchain interaction energy
- Example: Interchain Interaction Energy
- intramolecular interactions
- Intramolecular Interactions
- j-coupling restraints
- Scalar J-Coupling Restraints
- Langevin dynamics
- Simple Langevin Dynamics | Example: Run Langevin Dynamics
- Lattman's angles
- 3
 3 Matrices
3 Matrices
- learning parameters
- no title | Learning Atom-based Parameters
- Lee and Richards algorithm
- Accessible Surface Area
- Lennard-Jones potential
- Nonbonded Energy Terms | Van der Waals Function
- letters,uppercase and lowercase
- Notation
- lipids,topologies and parameters
- CHARMM “top_all22*" and “par_all22*"
- loops
- Example: A Simple Loop | Example: A Double Loop | Example: Switch Control to
- Luzzati plot
- Luzzati Plot
- manipulating coordinates
- Coordinate Statement
- manipulation,diffraction data
- Manipulating Reflection Data
- Mathematica
- Mathematica Interface
- matrices
- 3
 3 Matrices
3 Matrices
- Maxwellian distribution function
- Syntax
- messages
- Set Statement
- metrix matrix distance geometry
- Distance Geometry
- metrization
- Metrization
- minimization
- Energy Minimization
- minimization,conjugate gradient
- Conjugate Gradient Minimization
- minimization,rigid-body
- Rigid-Body Minimization
- mirror images
- Test for the Correct
- molecular dynamics
- Molecular Dynamics
- molecular dynamics,angular distribution
- Angular Distribution Functions
- molecular dynamics,average displacement
- Average Coordinates and Fluctuations
- molecular dynamics,covariance
- Covariance Analysis
- molecular dynamics,density
- Density Analysis
- molecular dynamics,finite difference approximation
- Finite Difference Approximation
- molecular dynamics,merging files
- Merging Trajectories
- molecular dynamics,orienting trajectory frames
- Syntax
- molecular dynamics,pick properties
- Picking Properties for Trajectories
- molecular dynamics,power spectrum
- Power Spectrum Analysis
- molecular dynamics,radial distribution function
- Radial Distribution Functions
- molecular dynamics,restarts
- Dynamics Restarts
- molecular dynamics,simulation
- Example: Run a Standard | Example: Run a Molecular | Example: Run a Slow-cooling
- molecular dynamics,slow-cooling
- Example: Run a Slow-cooling
- molecular dynamics,temperature control
- Temperature Control
- molecular dynamics,temperature control,Langevin dynamics
- Langevin Dynamics
- molecular dynamics,temperature control,temperature coupling
- Temperature Coupling
- molecular dynamics,temperature control,velocity rescaling
- Velocity Rescaling
- molecular dynamics,time correlation
- Time Correlation Analysis
- molecular dynamics,trajectories
- Management of Trajectories
- molecular dynamics,trajectory analysis
- Analysis of Trajectories
- molecular dynamics,velocity assignment
- Velocity Assignment
- molecular replacement
- Molecular Replacement
- molecular replacement,cluster analysis
- The Rotation Function Listing
- molecular replacement,cluster criteria
- Rotation Search
- molecular replacement,comparing rotation matrices
- Comparing Orientations of Molecules
- molecular replacement,contour plots
- A Mathematica Script File
- molecular replacement,cross rotation function,PC-refinement
- PC-Refinement of the Highest | Analysis of the PC-refinement
- molecular replacement,cross-rotation function
- Cross-Rotation Function with the
- molecular replacement,direct rotation function
- A “Direct" Rotation Function
- molecular replacement,elbow angle modification
- Modification of the Elbow
- molecular replacement,packing function
- A Packing Function
- molecular replacement,rigid-body refinement
- Rigid-Body Refinement
- molecular replacement,rotation search
- Rotation Search
- molecular replacement,self-rotation function
- Self-rotation Function
- molecular replacement,translation function,asymmetric unit
- Translation Search
- molecular replacement,translation function,finding second molecule
- Translation Function for Molecule
- molecular replacement,translation function,relative
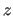 position
position
- Combined Translation Function to
- molecular replacement,translation function,using
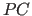 -refined model
-refined model
- Translation Function for Molecule
- molecular replacement,translation search
- Translation Search
- molecular structure file
- Example: How to Read
- molecular structure,append
- Example: Append Two Molecular
- molecular structure,deletion
- Deleting Atoms
- molecular structure,disulfide bridge
- Example: Incorporation of Disulfide
- molecular structure,duplicating
- Duplicating the Molecular Structure
- molecular structure,examples
- Examples for Molecular Structure
- molecular structure,generation
- Generating the Molecular Structure
- molecular structure,histidine protonation
- Example: Modification of the
- molecular structure,incomplete polypeptide chain
- Example: A Polypeptide Chain
- molecular structure,nucleic acid
- A Nucleic Acid Structure
- molecular structure,patching
- Syntax | Patching the Molecular Structure
- molecular structure,polypeptide chain
- Example: A Polypeptide Chain
- molecular structure,protein
- A Standard Protein Structure
- molecular structure,protein,unusual geometries
- How to Set Up
- molecular structure,reading
- Structure Statement
- molecular structure,sequence
- Syntax
- molecular structure,solvent and solute
- Example: Solvation of a
- molecular structure,unknown atoms
- What to Do about
- molecular structure,viruses or multimers
- Virus Structures or Structures
- molecular structure,water
- Example: Water Molecules
- molecular structure,writing
- Writing a Molecular Structure
- NCS
- Non-crystallographic Symmetry | Strict NCS
- Newton's equations of motion
- Molecular Dynamics
- NMR relaxation refinement,
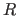 value
value
- Assessing the Quality of
- NMR relaxation refinement,against NOESY intensities
- Refinement against NOESY Intensities
- NMR relaxation refinement,cutoffs
- Cutoffs
- NMR relaxation refinement,including solvent spectra
- Simultaneous Refinement with H
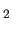 O
O
- NMR relaxation refinement,input of experimental data
- Input of the Experimental
- NMR relaxation refinement,optimal correlation time
- Grid Search for Optimal
- NMR relaxation refinement,quality assessment
- Assessing the Quality of
- NMR structure determination
- NMR Structure Determination
- NMR structure determination, topologies and parameters
- Files “parallhdg.dna" and “topallhdg.dna"
- NMR structure determination,acceptance of refined structures
- Acceptance of Refined NMR
- NMR structure determination,average structure
- Average Structure and Rmsds
- NMR structure determination,enantiomer selection
- Test for the Correct
- NMR structure determination,rms differences
- Average Structure and Rmsds | Pairwise Rmsds
- NMR structure determination,simulated annealing
- Ab Initio SA Starting | Random Simulated Annealing | Simulated Annealing Refinement
- NMR structure determination,substructure distance geometry
- Distance Geometry
- NMR structure determination,template coordinate set
- Template Structure
- NMR structure determination,time averaging
- Time-Average Refinement
- NMR structure refinement, topologies and parameters
- Files “parallhdg.dna" and “topallhdg.dna"
- NMR, relaxation matrix refinement
- NMR Back-calculation Refinement
- NOE
- Distance Restraints
- NOE back-calculation
- NMR Back-calculation Refinement
- NOE back-calculation,BPTI example
- Prediction of a NOESY
- NOE back-calculation,gradient expression
- Analytical Expression for the
- NOE back-calculation,leakage
- The Relaxation Matrix
- NOE back-calculation,unresolved chemical shifts
- Syntax
- NOE distances, restraints
- Distance Restraints
- NOE distances,ambiguous
- Expressing ambiguous restraints using
- NOE distances,averaging methods
- Choice of Averaging
- NOE distances,distance restraints file
- Setup of Distance Restraints
- NOE distances,pseudoatoms
- Pseudoatoms
- NOE distances,restraining functions
- Choice of Restraining Functions
- NOE, 3D
- 3D NOE-NOE Example
- NOE,time-average
- Refinement Using Time-Averaged Distance
- non-crystallographic symmetry
- Non-crystallographic Symmetry
- non-crystallographic symmetry,Bricogne conventions
- Strict NCS
- non-crystallographic symmetry,restrained
- Non-crystallographic Symmetry
- non-crystallographic symmetry,restraints
- NCS Restraints
- non-crystallographic symmetry,skew frame
- Strict NCS
- non-crystallographic symmetry,strictly identical
- Non-crystallographic Symmetry | Strict NCS
- nonbonded distances
- Analysis of the Nonbonded
- nonbonded energy terms
- Nonbonded Energy Terms | Analysis of the Nonbonded
- nonbonded energy terms,truncation
- Nonbonded Energy Terms
- nonbonded interaction, 1-4
- Syntax | Syntax | Syntax | Syntax | Files “parhcsdx.pro" and “tophcsdx.pro" | Intramolecular Interactions
- nonbonded parameters
- Syntax
- nucleic acids,molecular structure
- A Nucleic Acid Structure
- nucleic acids,topologies and parameters
- CHARMM “top_all22*" and “par_all22*" | CHARMM “toph11.dna" and “param11.dna" | Files “topnah1e.dna" and “parnah1e.dna" | Files “parallhdg.dna" and “topallhdg.dna"
- numerical precision
- Set Statement
- occupancy refinement
- Grouped B-Factor and Occupancy
- one-bond coupling restraints
- One-Bond Coupling Restraints
- ONEBond-statement
- Syntax
- ORIEnt-statement
- Syntax
- orthogonalized coordinates
- Examples | Orthogonalization Convention
- packing function
- Crystallographic Target Functions | A Packing Function
- paramagnetic cross-correlation rate restraint
- Paramagnetic Cross-Correlation Rate Restraint
- paramagnetic orientation restraint
- Paramagnetic Orientation Restraint
- paramagnetic pseudocontact shift restraint
- Paramagnetic Pseudocontact Shift Restraint
- paramagnetic relaxation enhancement, distance calculation
- Distance Calculation from Paramagnetic
- paramagnetic relaxation enhancement, restraints
- Paramagnetic Relaxation Enhancement Restraints
- paramagnetic residual dipolar coupling restraints
- Paramagnetic Residual Dipolar Coupling
- parameter and topology files
- Topology and Parameter Files
- parameter and topology files, lipids
- CHARMM “top_all22*" and “par_all22*"
- parameter and topology files, water
- Files “toph19.sol" and “param19.sol"
- parameter and topology files,carbohydrates
- Files “toph3.cho" and “param3.cho"
- parameter and topology files,chromophores
- Files “toph19.chromo" and “param19.chromo"
- parameter and topology files,crystallographic refinement
- Files “parhcsdx.pro" and “tophcsdx.pro"
- parameter and topology files,nucleic acids
- CHARMM “top_all22*" and “par_all22*" | CHARMM “toph11.dna" and “param11.dna" | Files “topnah1e.dna" and “parnah1e.dna" | Files “parallhdg.dna" and “topallhdg.dna"
- parameter and topology files,proteins
- CHARMM “top_all22*" and “par_all22*" | CHARMM “toph19.pro" and “param19.pro" | AMBER/OPLS “tophopls.pro", “parhopls.pro" Files | Files “parhcsdx.pro" and “tophcsdx.pro" | Files “parallhdg.pro" and “topallhdg.pro"
- parameter,writing
- Writing a Parameter File
- parameters
- Parameter Statement
- parameters, atom-based
- Parameter Statement | Examples of Atom-based Modifications
- parameters, hydrogen-bonding
- Syntax
- parameters, nonbonded
- Syntax
- parameters, type-based
- Parameter Statement
- parameters,learning
- no title | Learning Atom-based Parameters
- parameters,reducing
- Reducing to Type-based Parameters
- parsing
- X-PLOR Language
- partial energy terms
- Symbols
- PATCh statement
- Topology Statement
- Patterson correlation (
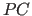 )
)
refinement - Molecular Replacement
- PCSA restraint
- Pseudo Chemical Shift Anisotropy
- PCSA-statement
- Syntax
- PDB format
- Syntax
- phase difference
- Syntax of the Xrefin
- phase restraints
- Crystallographic Target Functions
- pick energy
- Syntax of the Pick
- pick geometry
- Syntax of the Pick
- planarity
- Conformational Energy Terms
- PMAG-statement
- Syntax
- Powell minimization
- Conjugate Gradient Minimization
- print energy
- Syntax of the Print
- print geometry
- Syntax of the Print
- proteins,molecular structure
- A Standard Protein Structure
- proteins,topologies and parameters
- CHARMM “top_all22*" and “par_all22*" | CHARMM “toph19.pro" and “param19.pro" | AMBER/OPLS “tophopls.pro", “parhopls.pro" Files | Files “parhcsdx.pro" and “tophcsdx.pro" | Files “parallhdg.pro" and “topallhdg.pro"
- proton chemical shift
- Proton Chemical Shift Restraints
- PROTonshifts statement
- Syntax
- pseudo chemical shift anisotropy restraint
- Pseudo Chemical Shift Anisotropy
- pseudoatoms
- Pseudoatoms | Pseudoatoms | NMR Structure Determination
- quanta
- Binary Trajectory Files
- QUANTA trajectory format
- Binary Trajectory Files
- quaternions
- 3
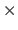 3 Matrices
| Initialization
3 Matrices
| Initialization
- R value
- Crystallographic Target Functions | Check of Data and
- R value, distribution
- Syntax of the Xrefin
- R value,free
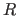 value
value
- Cross-validation: The Free
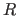
- R value,Luzzati plot
- Luzzati Plot
- R value,NMR structure
- Assessing the Quality of
| Calculation of Different
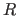
- radius of gyration
- Syntax | Scaling
- radius of gyration restraint
- Radius of Gyration Restraints
- Ramachandran plot
- Ramachandran Plot
- RAMAchandran-statement
- Syntax
- random-number function
- Syntax
- random-number seed
- Set Statement
- RDC tensor axis atoms
- Requirements
- RDCs
- Residual Dipolar Couplings
- reading,coordinates
- Coordinate Statement
- reading,distance restraints
- Setup of Distance Restraints
- reading,molecular structure
- Structure Statement
- reading,trajectory
- Reading Trajectories
- reducing parameters
- Reducing to Type-based Parameters
- reflection files
- Reflection Files | Example: A Crystallographic Reflection
- reflection files,merging files
- Example: Merging Crystallographic Reflection
- reflections,manipulation
- Manipulating Reflection Data
- reflections,writing
- Syntax
- relaxation energy term
- The
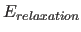 Energy Term
Energy Term
- relaxation matrix
- The Relaxation Matrix
- relaxation matrix refinement, NMR
- NMR Back-calculation Refinement
- remarks
- Control Statements
- repel energy term
- Nonbonded Energy Terms
- residual dipolar couplings
- Residual Dipolar Couplings
- residue-residue positional restraints, database
- Residue-Residue Position/Orientation Database Restraint
- residues
- Syntax
- restraints, dihedral angle
- Dihedral Angle Restraints
- restraints, harmonic plane
- Plane Restraints
- restraints, harmonic point
- Point Restraints
- restraints,coordinate
- Coordinate Restraints
- restraints,dihedral
- Dihedral Angle Restraints
- restraints,distance
- Distance Restraints
- restraints,distance symmetry
- Distance Symmetry Restraints
- restraints,harmonic coordinate
- Harmonic Coordinate Restraints
- restraints,planarity
- Planarity Restraints
- rigid-body minimization
- Rigid-Body Minimization
- rigid-body molecular dynamics
- Rigid-Body Coordinate Space
- rigid-body refinement
- Rigid-Body Refinement
- ring pucker
- Syntax of the Pick | Syntax of the Pick
- ring pucker,nucleic acids
- CHARMM “toph11.dna" and “param11.dna"
- rms deviation
- Syntax
- rmsd
- Rms Differences between Coordinates | Average Structure and Rmsds | Pairwise Rmsds
- rotation function,list file
- The Rotation Function Listing
- rotation function,output file
- The Rotation Function Output
- rotation matrix (3
 3)
3)
- 3
 3 Matrices
3 Matrices
- rotation search
- Rotation Search
- SANI statement
- Syntax - SANI
- scalar j-coupling
- Scalar J-Coupling Restraints
- scaling of diffraction data
- Crystallographic Target Functions | Syntax
- segment statement
- Topology Statement
- selection
- Atom Selection
- SHAKE
- Fixing Distances
- simulated annealing,ab initio from template
- Ab Initio SA Starting
- simulated annealing,crystallographic refinement
- Crystallographic Refinement by Simulated
- simulated annealing,NMR refinement
- Simulated Annealing Refinement
- simulated annealing,preparation
- Crystallographic Refinement by Simulated
- simulated annealing,random coordinates
- Random Simulated Annealing
- simulated annealing,slow-cooling
- Example: Run a Slow-cooling | Crystallographic Refinement by Simulated | Simulated Annealing Refinement
- slow-cooling molecular dynamics
- Example: Run a Slow-cooling
- slow-cooling,crystallographic refinement
- Crystallographic Refinement by Simulated
- solvation
- Example: Solvation of a
- solvent boundary forces
- Deformable Boundary Forces
- solvent mask
- Bulk Solvent Mask | Example: Use of a
- solvent screening
- Electrostatic Function
- special positions
- Special Positions
- structure factor,normalized
- Crystallographic Target Functions
- structure factor,partial
- Crystallographic Target Functions | Partial Structure Factors
- structure factor,solvent
- Example: Use of a
- structure factors
- Crystallographic Target Functions
- structure generation,nucleic acid
- A Nucleic Acid Structure
- structure generation,protein
- A Standard Protein Structure
- structure generation,protein,with cofactor or substrate
- A Protein Structure with
- structure generation,protein,with metal cluster
- A Protein Structure with
- structure generation,protein,with water or ligands
- A Protein Structure with
- structure generation,viruses or multimers
- Virus Structures or Structures
- structure,generation
- Generating the Molecular Structure
- surface statement
- Accessible Surface Area
- symbols
- Symbols
- symmetry interactions,crystallographic
- Crystallographic Symmetry Interactions
- symmetry interactions,non-crystallographic
- Non-crystallographic Symmetry Interactions
- symmetry operators
- Crystallographic Symmetry Interactions | Syntax | Crystallographic Target Functions | Strict NCS
- symmetry,distance restraints
- Distance Symmetry Restraints
- tag
- Syntax | Conformation vs. Residue Number
- TENSOr statement
- Syntax - TENSOr
- time-averaged distance restraints
- Refinement Using Time-Averaged Distance
- topology
- Topology Statement
- trajectory statement
- Trajectory Statement
- trajectory,analysis
- Analysis of Trajectories
- trajectory,analysis,angular distribution
- Angular Distribution Functions
- trajectory,analysis,average
- Average Coordinates and Fluctuations
- trajectory,analysis,covariance
- Covariance Analysis
- trajectory,analysis,density
- Density Analysis
- trajectory,analysis,pick properties
- Picking Properties for Trajectories
- trajectory,analysis,power spectrum
- Power Spectrum Analysis
- trajectory,analysis,radial distribution function
- Radial Distribution Functions
- trajectory,analysis,time correlation
- Time Correlation Analysis
- trajectory,error messages
- Diagnostic Error Messages
- trajectory,merging coordinate files
- Example
- trajectory,merging files
- Merging Trajectories
- trajectory,reading
- Reading Trajectories
- trajectory,writing
- Writing Trajectories
- translation function,output file
- The Translation Function Output
- translation function,peak list
- The Translation Function Listing
- translation search
- Translation Search
- type-based parameters
- Parameter Statement
- unit cell constants
- Crystallographic Symmetry Interactions
- unknown coordinates
- What to Do about
- Van der Waals radii, parameters
- Van der Waals radii
- van der Waals radius
- Van der Waals Function
- van der Waals radius, in NMR protocols
- NMR Structure Determination
- VEANgle statement
- Syntax- VEANngle
- vector
- Vector Statement
- vectors (3d)
- Three-dimensional Vectors
- Verlet method
- Finite Difference Approximation
- viruses,molecular structure
- Virus Structures or Structures
- water
- Example: Water Molecules | Example: Solvation of a
- water hydrogens
- Building Hydrogen Positions
- water,topologies and parameters
- Files “toph19.sol" and “param19.sol"
- while
- Control Statements
- wildcards
- Wildcards
- Wilson plot
- Syntax of the Xrefin | Wilson Plot
- writing,coordinates
- Write Coordinate Statement
- writing,molecular structure
- Writing a Molecular Structure
- writing,reflections
- Syntax
- writing,trajectory
- Writing Trajectories
- X-PLOR,input
- X-PLOR Language
- X-PLOR,notation
- Notation
- XANGle-statement
- Syntax
- XCCR-statement
- Syntax
- XDIPolar statement
- Syntax - XDIPolar or
- XPCS-statement
- Syntax
- XRDCoupling-statement
- Syntax
- XT1D-statement
- Syntax
Xplor-NIH 2025-11-07
