| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Grid Search for Optimal Up: NMR Back-calculation Refinement Previous: Simultaneous Refinement with HO
Calculation of Different 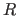 Values
Values
The following example shows a part of the output file, i.e., the calculated
calibration factor and the ![]() value.
The intensities are calculated from
the coordinates by the calibration statement, and the
optimal calibration factor is printed for each classification
separately. The
value.
The intensities are calculated from
the coordinates by the calibration statement, and the
optimal calibration factor is printed for each classification
separately. The ![]() value is printed first for each
classification
and then for each group broken down in intensity shells.
value is printed first for each
classification
and then for each group broken down in intensity shells.
RELAx> calibrate
CALIbrate> quality 0.33 automatic off reference all group class
CALIbrate> end
RELAX: intensities updated for calibration
class N(reference) N(all) calibration
---------------------------------------------
D300 0 23 .5220E+00
---------------------------------------------
RELAx>
RELAx> print threshold -0.1 end {List all deviations and calculate R.}
Class D300: R-factor: .6594E-01 R.m.s. difference: .8038E-01
RELAX: overall R-factor statistics
------------------------------------------------------
min. int. max. int. N R-factor r.m.s. diff
------------------------------------------------------
.2550E+01 .5100E+01 1 .4804E-01 .6302E-01
.1275E+01 .2550E+01 4 .5346E-01 .8720E-01
.6375E+00 .1275E+01 4 .4047E-01 .4735E-01
.3187E+00 .6375E+00 9 .6331E-01 .7320E-01
.1594E+00 .3187E+00 5 .1292E+00 .1204E+00
.7969E-01 .1594E+00 4 .5022E-01 .5105E-01
------------------------------------------------------
.0000E+00 .5100E+01 27 .6594E-01 .8038E-01
------------------------------------------------------
A local
relaxation
set echo off message off end
for $id in id (name ca) loop rfac
print
selection=(byres(id $id))
threshold 9999
end
end loop rfac
set echo on message on end
end
Different types of local ![]() values can be calculated using
the atom selection routine. For example, the
values can be calculated using
the atom selection routine. For example, the ![]() value for
the region around each residue can be calculated with the
around option of the atom selection command (see Section
2.15).
value for
the region around each residue can be calculated with the
around option of the atom selection command (see Section
2.15).
Xplor-NIH 2025-11-07
