| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Self-rotation Function Up: Molecular Replacement Previous: A Mathematica Script File
Generalized Molecular Replacement
The following sections show how the 26-10 Fab fragment complexed with digoxin was solved by generalized molecular replacement (Brünger, 1991c). The space group of the 26-10 Fab/digoxin crystals is- Self-rotation function.
- Modification of the elbow angle of a known Fab structure.
- Cross-rotation function with the modified Fab structure.
- Filtering the rotation function by
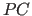 -refinement.
-refinement.
- Analysis of the
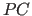 -refinement.
-refinement.
- Translation function for molecule A, using the
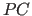 -refined model.
-refined model.
- Translation function for molecule B.
- Combined translation function to determine the relative position between A,B.
- Rigid-body refinement.
Subsections
- Self-rotation Function
- Modification of the Elbow Angle of a Known Fab Structure
- Cross-Rotation Function with the Modified Fab Structure
- PC-Refinement of the Highest Peaks of the Cross-Rotation Function
- Analysis of the PC-refinement
- Translation Function for Molecule A Using the PC-refined Model
- Translation Function for Molecule B
- Combined Translation Function to Determine the Relative Position between A and B
- Rigid-Body Refinement

