| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Abbreviations Up: XPLOR Interface Manual Previous: List of Application Statements
Bibliography
- Bax, A., Kontaxis, G. and Tjandra, N. (2001)
- “Dipolar couplings in macromolecular structure determination”,
Methods Enzymol. 339, 127-174.
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, N.F., DiNola, A., and Haak, J.R. (1984).
- “Molecular dynamics
with coupling to an external bath", J. Chem. Phys.
81, 3684-3690.
- Bermejo G.A. and Llinás M. (2008).
- “Deuterated Protein Folds
Obtained Directly from Unassigned Nuclear Overhauser Effect Data",
J. Am. Chem. Soc. ASAP Article 10.1021/ja074836e.
- Banci, L., Bertini, I., Cavallaro, G., Giachetti, A., Luchinat, C., and Parigi, G. (2004) ;
- SPMldquo;Paramagnetism-based restraints for Xplor-NIH",
J. Biomol. NMR 28, 249-261.
- Bernstein, R., Ross, A., Cieslar, C., and Holak, T. A. (1993).
-
“A Practical Approach to Calculations of Biomolecular Structures from
Homonuclear Three-Dimensional NOE-NOE Spectra",
J. Magn. Reson. B 101, 185-188.
- Bricogne, G. (1976).
- “Methods and Programs for Direct-Space Exploitation
of Geometric Redundancies", Acta Cryst. A32,
832-847.
- Brooks, B., Bruccoleri, R., Olafson, B., States, D., Swaminathan, S., and Karplus, M. (1983).
-
“CHARMM: A Program for Macromolecular Energy,
Minimization, and Molecular Dynamics Calculations",
J. Comp. Chem. 4, 187-217.
- Brooks, C. L. III and Karplus M. (1983).
-
“Deformable stochastic boundaries in molecular dynamics”,
J. Chem. Phys. 79, 6312-6325.
- Brünger, A.T. (1988).
- “Crystallographic Refinement by Simulated
Annealing: Application to a 2.8 Å Resolution Structure of
Aspartate Aminotransferase", J. Mol. Biol. 203,
803-816.
- Brünger, A.T. (1989).
- “A Memory-Efficient Fast Fourier Transformation
Algorithm for Crystallographic Refinement on Supercomputers",
Acta Cryst. A45, 42-50.
- Brünger, A.T. (1990).
- “Extension of Molecular Replacement: A
New Search Strategy based on Patterson
Correlation Refinement", Acta Cryst.
A46, 46-57.
- Brünger, A.T. (1991a).
- “Crystallographic Phasing and Refinement of
Macromolecules",
Current Opinion in Structural Biology 1, 1016-1022.
- Brünger, A.T. (1991b).
- “Simulated Annealing in Crystallography",
Ann. Rev. Phys. Chem. 42, 197-223.
- Brünger, A.T. (1991c).
- “Solution of a Fab (26-10) /Digoxin Complex by
Generalized Molecular Replacement", Acta Cryst. A47,
195-204.
- Brünger, A.T. (1992).
- “The Free
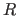 value: a Novel Statistical Quantity for
Assessing the Accuracy of Crystal Structures", Nature
355, 472-474.
value: a Novel Statistical Quantity for
Assessing the Accuracy of Crystal Structures", Nature
355, 472-474.
- Brünger, A.T. (1993).
- “Assessment of Phase Accuracy by Cross
Validation: The Free
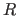 Value. Methods and Applications",
Acta Cryst. D 49, 24-36.
Value. Methods and Applications",
Acta Cryst. D 49, 24-36.
- Brünger, A.T., Brooks, C.L., and Karplus, M. (1984).
- “Stochastic
Boundary-Conditions for Molecular-Dynamics Simulations of
ST2 Water", Chem. Phys. Lett. 105, 495-500.
- Brünger, A.T., Clore, G.M., Gronenborn, A.M., and Karplus, M. (1986).
-
“Three-dimensional structures of proteins determined by molecular dynamics
with interproton distance restraints: Application to crambin",
Proc. Natl. Acad. Sci. USA 83, 3801-3805.
- Brünger, A.T., Clore, G.M., Gronenborn, A.M., and Karplus, M. (1987).
-
“Solution
conformations of a human growth hormone releasing factor: comparison of
the restrained molecular dynamics and distance geometry methods for a system
without long-range distance data",
Protein Engineering 1, 399-406.
- Brünger, A.T. and Karplus, M. (1988).
- “Polar hydrogen positions in
proteins: Empirical energy function placement and neutron
diffraction comparison", Proteins 4,148-156.
- Brünger, A.T. and Karplus, M. (1991).
- “Molecular Dynamics
Simulations with
Experimental Restraints", Accounts Chemical Research
24 , 54-61.
- Brünger, A.T., Karplus, M., and Petsko, G.A. (1989).
- “Crystallographic
Refinement by Simulated Annealing: Application to a 1.5 Å Resolution Structure of Crambin", Acta Cryst. A45,
50-61.
- Brünger, A. T., Krukowski, A., and Erickson, J. (1990).
- “Slow-Cooling
Protocols for Crystallographic Refinement by Simulated Annealing",
Acta Cryst. A46, 585-593.
- Brünger, A.T., Kuriyan, J., and Karplus, M. (1987).
-
“Crystallographic
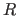 Factor Refinement by Molecular Dynamics",
Science 235, 458-460.
Factor Refinement by Molecular Dynamics",
Science 235, 458-460.
- Brünger, A.T., Leahy, D.J., Hynes, T.R., and Fox, R.O. (1991).
-
“The 2.9 Å Resolution Structure of an
Anti-Dinitrophenyl-Spin-Label
Monoclonal Antibody Fab Fragment with Bound Hapten",
J. Mol. Biol. 221 , 239-256.
- Burkert, U. and Allinger, N.L. (1982).
- In: Molecular Mechanics,
ACS Monograph 177, American Chemical Society, Washington.
- Clore, G.M., Appella, E., Yamada, M., Matsushima, K., and Gronenborn, A.M. (1990).
-
“Three-Dimensional Structure of Interleukin 8 in Solution",
Biochemistry 29, 1689-1696.
- Clore, G.M. and Gronenborn, A.M. (1989).
-
“Determination of three-dimensional
structures of proteins and nucleic acids in solution by nuclear magnetic
resonance spectroscopy",
CRC Critical Reviews in Biochemistry
24, 479-564.
- Clore, G.M. and Gronenborn, A.M. (1991).
-
“Structures of Larger Proteins in Solution:
Three- and Four-Dimensional Heteronuclear NMR Spectroscopy",
Science 252, 1390-1399.
- Clore, G.M., Gronenborn, A.M., and Tjandra, N. (1998).
- “Direct
Structure Refinement against Residual Dipolar Couplings in the Presence
of Rhombicity of Unknown Magnitude",
J. Magn. Reson. 131, 159-162.
- Clore, G.M., Gronenborn, A.M., and Bax, A. (1998).
- “A robust method
for determining the magnitude of the fully asymmetric alignment tensor
of oriented macromolecules in the absence of structural information",
J. Magn. Reson. 133, 216-221.
- Clore, G.M., Gronenborn, A.M., Szabo, A., and Tjandra, N. (1998).
-
“Determining the magnitude of the fully asymmetric diffusion tensor
from heteronuclear relaxation data in the absence of structural
information",
J. Am. Chem. Soc.
120, 4889-4890.
- Clore, G.M., Starich, M.R., Bewley, C.A., Cai., M. and Kuszewski, J. (1999).
- “Impact of residual dipolar couplings on the accuracy of NMR
structures determined from a minimal number of NOE restraints",
J. Am. Chem. Soc. 121, 6513-6514.
- Clore, G.M. and Bewley, C.A. (2002).
- “Using conjoined rigid body/
torsion angle simulated annealing to determine the relative orientation
of covalently linked protein domains from dipolar couplings",
J. Magn. Reson. 154, 329-335.
- Clore, G.M. and Kuszewski, J. (2002).
- “
 rotamer populations and
angles of mobile surface side chains are accurately predicted by a
torsion angle database potential of mean force",
J. Am. Chem. Soc. 124, 2866-2867.
rotamer populations and
angles of mobile surface side chains are accurately predicted by a
torsion angle database potential of mean force",
J. Am. Chem. Soc. 124, 2866-2867.
- Clore, G.M. and Kuszewski, J. (2003).
- “Improving the accuracy of NMR
structures of RNA by means of conformational database potentials of
mean force as assessed by complete dipolar coupling cross-validation",
J. Am. Chem. Soc. 125, 1518-1525.
- Clore, G.M. and Schwieters, C.D. (2003).
- “Docking of protein-protein
complexes on the basis of highly ambiguous intermolecular distance
restraints derived from
 HN/
HN/ N chemical shift mapping and backbone
N chemical shift mapping and backbone
 N-
N- H residual dipolar couplings using conjoined rigid body/torsion
angle dynamics",
J. Am. Chem. Soc.
125, 2902-2912.
H residual dipolar couplings using conjoined rigid body/torsion
angle dynamics",
J. Am. Chem. Soc.
125, 2902-2912.
- Cremer, D. and Pople, J.A. (1975).
- “General Definition of
Ring Puckering Coordinates", J. Am. Chem. Soc. 97,
1354-1358.
- Crippen, G. and Havel T. (1988).
- Distance
Geometry and Molecular Conformation. Research Studies Press:
Taunton, Somerset, England.
- Denny, R.C., Shotton, M.W., and Forsyth, V.T. (1997).
- “X-PLOR for
Polycrystalline Fibre Diffraction", Fibre Diffr. Rev. 6, 30-33.
- Donaldson, L.W., Skrynnikov, N.R., Choy, W.-Y., Muhandiram, D.R., Sarkar, B., Forman-Kay, J.D., and Kay, L.E. (2001).
- “Structural
Characterization of Proteins with an Attached ATCUN Motif by
Paramagnetic Relaxation Enhancement NMR Spectroscopy",
J. Am. Chem. Soc. 123, 9843-9847.
- Engh, R.A. and Huber, R. (1991).
- “Accurate Bond and
- Ernst, R.R., Bodenhausen, G., and Wokaun, A. (1987).
-
Principles of Nuclear Magnetic Resonance in One and
Two Dimensions. Oxford: Clarendon Press.
- Garrett, D.S., Kuszewski, J., Hancock, T.J., Lodi, P.J., Vuister, G.W., Gronenborn, A.M., and Clore, G.M. (1994).
- “The Impact of Direct
Refinement against Three-Bond HN-CaH Coupling Constants on Protein
Structure Determination by NMR",
J. Magn. Reson. B 104, 99-103.
- Goldstein, H. (1980).
- Classical Mechanics. New York: Addison-Wesley,
chap. 4.
- Grishaev, A., and Bax, A. (2004).
- “An empirical backbone-backbone
potential in proteins and its application to NMR structure refinement
and validation", J. Am. Chem. Soc. 126, 7281-7292.
- Grishaev, A., Wu, J., Trewhella, J., and Bax, A. (2005).
- “Refinement of
multidomain protein structures by combination of solution small-angle
X-ray scattering and NMR data”, J. Am. Chem. Soc. 127,
16621-16628.
- Grishaev, A., Ying, J., and Bax, A. (2006).
- “Pseudo-CSA restraints
for NMR refinement of nucleic acid structure"
J. Am. Chem. Soc. 128 10010-10011.
- Gronenborn, A.M., Filpula, D.R., Essig, N.Z., Achari, A., Whitlow, M., Wingfield, P.T., and Clore, G.M. (1991).
-
“The immunoglobulin binding domain of streptococcal protein G
has a novel and highly stable polypeptide fold",
Science 253, 657-661.
- Ha, S.N., Giammona, A., Field, M. and Brady, J.W. (1988).
-
“A Revised Potential Energy Surface for Molecular Mechanics
Studies of Carbohydrates", Carbohydrate
Res. 180, 207-221.
- Habazettl, J., Cieslar, C., Oschkinat, H., and Holak, T.A. (1990).
- “H-1-NMR Assignments of Side-Chain Conformations in Proteins
Using a High-Dimensional Potential in the Simulated Annealing
Calculations", FEBS Lett. 268, 141-145.
- Habazettl, J., Ross, A., Oschkinat, H. and Holak, T. A. (1992a).
-
“Secondary NOE pathways in 2D NOESY spectra of proteins estimated from
homonuclear three-dimensional NOE-NOE nuclear magnetic resonance
spectroscopy",
J. Magn. Reson. 97, 281-294.
- Habazettl, J., Schleicher, M., Otlewski, J., and Holak, T. A. (1992b).
-
“Homonuclear Three-Dimensional NOE-NOE NMR Spectra for Structure
Determination of Proteins in Solution",
J. Mol. Biol., 228, 156-169.
- Hahn, T., ed. (1987).
- International Tables for
Crystallography, Vol. A. , D. Reidel Publishing Company.
-
“A distance geometry program for determining the
structures of small proteins and other macromolecules from nuclear
magnetic resonance measurements of intramolecular
 H
H H proximities
in solution"
Bull. Math. Bio. 46, 673-698.
H proximities
in solution"
Bull. Math. Bio. 46, 673-698.
- Head-Gordon, T. and Brooks, C.L. (1991).
- “Virtual Rigid Body Dynamics",
Biopolymers 31, 77-100.
- Hendrickson, W.A. (1985).
- “Stereochemically
Restrained Refinement of Macromolecular Structures",
Meth. Enzymol. 115, 252-270.
- Hendrickson, W.A. and Ward, K.B. (1976).
- “A Packing Function for
Delimiting the Allowable Locations of Crystallized Macromolecules",
Acta Cryst. A32, 778-780.
- Hirshfeld, F.L. (1968).
- “Symmetry in the Generation of Trial Structures",
Acta Cryst. A 24, 301-311.
- Hodel, A., Kim, S.-H., and Brünger, A.T. (1992).
-
“Model Bias in Macromolecular Crystal Structures", Acta Cryst. A,
48, 851-858.
- Huber, R. (1985).
- “Experience with the application of Patterson
search techniques", Molecular Replacement, (Machin, P.A., comp.),
Proceedings
of the Daresbury Study Weekend, Science and Engineering Research
Council, The Librarian, Daresbury Laboratory, Daresbury, United Kingdom,
58-61.
- James, T.L., Gochin, M., Kerwood, D.J., Schmitz, U., and Thomas, P.D. (1991).
- In: Computational Aspects of the Study of Biological
Macromolecules by NMR, (J.C. Hoch, ed.). New York: Plenum Press.
- Jorgensen, W., Chandrasekar, J., Madura, J., Impey, R., and Klein, M. (1983).
-
“Comparison of simple potential functions for simulating liquid water",
J. Chem. Phys. 79, 926-935.
- Jorgensen, W. and Tirado-Rives, J. (1988).
- “The OPLS potential function
for
proteins, energy minimization for crystals of cyclic peptides and crambin",
J. Am. Chem. Soc. 110, 1657-1666.
- Kabsch, W. (1976).
- “A Solution for the Best Rotation to
Relate Two Sets of Vectors.", Acta Cryst. A32, 922.
- Kaptein, R., Zuiderweg, E.R.P., Scheek, R.M., Boelens, R., and van Gunsteren, W.F. (1985).
- “A protein structure from NMR data. lac Repressor
headpiece", J. Mol. Biol.
182, 179-182.
- Karplus, M. and Petsko, G.A. (1990).
- “Molecular-Dynamics Simulations
in Biology", Nature 347, 631-39.
- Keepers, J. W. and James, T. L. (1984).
- “A Theoretical
Study of Distance Determinations from NMR. Two-Dimensional
Nuclear Overhauser Effect Spectra",
J. Magn. Reson. 57, 404-426.
- Kirkpatrick, S., Gelatt, C.D.Jr., Vecchi, M.P. (1983).
- “Optimization by
Simulated Annealing", Science 220, 671-680.
- Kuszewski, J., Nilges, M., and Brünger A.T. (1992).
- “Sampling and
efficiency of metric matrix distance geometry: A novel “partial"
metrization algorithm", J. Biomol. NMR
2, 33-56.
- Kuszewski, J., Qin, J., Gronenborn, A.M., and Clore, G.M. (1995).
-
“The Impact of Direct Refinement against
 C
C and
and  C
C Chemical Shifts on Protein
Structure Determination by NMR",
J. Magn. Reson. B 106, 92-96.
Chemical Shifts on Protein
Structure Determination by NMR",
J. Magn. Reson. B 106, 92-96.
- Kuszewski, J., Gronenborn, A.M., and Clore, G.M. (1995).
- “The Impact
of Direct Refinement against Proton Chemical Shifts on Protein
Structure Determination by NMR",
J. Magn. Reson. B 107, 293-297.
- Kuszewski, J., Gronenborn, A.M., and Clore, G.M. (1996a).
- “A
Potential Involving Multiple Proton Chemical-Shift Restraints for
Nonstereospecifically Assigned Methyl and Methylene Protons",
J. Magn. Reson. B 112, 79-81.
- Kuszewski, J., Gronenborn, A.M., and Clore, G.M. (1996b).
- “Improving
the quality of NMR and crystallographic protein structures by means of
a conformational database potential derived from structure databases",
Protein Science 5, 1067-1080.
- Kuszewski, J., Gronenborn, A.M., and Clore, G.M. (1997).
-
“Improvements and extensions in the conformational database potential
for the refinement of NMR and X-ray structures of proteins and nucleic
acids",
J. Magn. Reson. 125, 171-177.
- Kuszewski, J., Gronenborn, A.M., and Clore, G.M. (1999).
- “Improving
the Packing and Accuracy of NMR Structures with a Pseudopotential for
the Radius of Gyration", J. Am.
Chem. Soc. 121, 1337-1338.
- Kuszewski, J., and Clore, G.M. (2000).
- “Sources of and solutions to
problems in the refinement of protein NMR structures against torsion
angle potentials of mean force",
J. Magn. Reson. 146, 249-254.
- Kuszewski, J., Schwieters, C., and Clore, G.M. (2001).
- “Improving
the Accuracy of NMR Structures of DNA by Means of a Database Potential
of Mean Force Describing Base-Base Positional Interactions",
J. Am. Chem. Soc. 123, 3903-3918.
- Lattman, E.E. (1985).
- “Use of the Rotation and Translation Functions",
Methods Enzymol. 115, 55-77.
- Lee, B. and Richards, F.M. (1971).
- “The Interpretation of
Protein Structures: Estimation of Static Accessibility",
J. Mol. Biol. 55, 379-400.
- Lifson, S. and Stern, P.S. (1982).
- “Born-Oppenheimer Energy Surfaces of
Similar Molecules - Interrelations Between Bond Lengths, Bond Angles,
and Frequencies of Normal Vibrations in Alkanes",
J. Chem. Phys. 77, 4542-4550.
- Lipari, G. and Szabo, A. (1982).
- “Model-Free Approach to the
Interpretation of Nuclear Magnetic-Resonance Relaxation in
Macromolecules .1. Theory and Range of Validity",
J. Am. Chem. Soc. 104, 4546-4559.
- Lipsitz, R.S. and Tjandra, N. (2001).
- “Carbonyl CSA restraints from
solution NMR for protein structure refinement",
J. Am. Chem. Soc. 123, 11065-11066.
- Lipsitz, R.S. and Tjandra, N. (2003).
- “
 N chemical sift anisotropy
in protein structure refinement and comparison with NH residual dipolar
couplings", J. Magn. Reson.
164, 171-176.
N chemical sift anisotropy
in protein structure refinement and comparison with NH residual dipolar
couplings", J. Magn. Reson.
164, 171-176.
- Lipsitz, R.S., Sharma, Y., Brooks, B.R., and Tjandra, N. (2002).
-
“Hydrogen bonding in high-resolution protein structures: a new method
to assess NMR protein geometry",
J. Am. Chem. Soc.
124, 10621-10626.
- Luzzati, P.V. (1952).
- “Traitement Statistique des Erreurs dans la
Determination des Structures Cristallines",
Acta Cryst 5, 802-810.
- Macura, S. and Ernst, R. R. (1980).
- “Elucidation of Cross
Relaxation in Liquids by Two-Dimensional N.M.R. Spectroscopy",
Mol. Phys. 41, 95-117.
- Main P. (1975).
- “Recent Developments in the MULTAN System - The
Use of Molecular Structure", In "Crystallographic Computing Techniques",
(Ahmed, F.R., Huml, K., Sedlácek, B., eds.), 98-103.
- Meiler, J., Blomberg, N., Nilges, M. and Griesinger, C. (2000).
- “A
new approach for applying residual dipolar couplings as restraints in
structure elucidation",
J. Biomol. NMR
16, 245-252.
- Morris, L., MacArthur, M.W., Hutchinson, E.G., and Thornton, J.M. (1992).
- Stereochemical Quality of Protein Structure
Coordinates, Proteins 12, 345-364.
- Naur, P. (1960).
- “Revised Report on Algorithmic Language Algol-60",
Computer J., 5, 349.
- Némethy, G., Pottie, M.S. and Scheraga, H.A. (1983)
- “Energy Parameters in Polypeptides. 9. Updating of Geometrical Parameters,
Nonbonded Interactions, and Hydrogen Bond Interactions for the
Naturally Occurring Amino Acids", J. Phys. Chem. 87, 1883-1887.
- Nilges, M. and Brünger, A.T. (1991),
- J. Cellular Biochem.,
Supplement 15G, Abstract, CG 422, 98.
- Nilges, M., Clore, G.M., and Gronenborn, A.M. (1988a).
-
“Determination of three-dimensional structures of
proteins from interproton distance data by dynamical simulated annealing
from a random array of atoms", FEBS Lett. 239, 129-136.
- Nilges, M., Clore, G.M., and Gronenborn, A.M. (1988b).
-
“Determination of
three-dimensional structures of proteins from interproton distance data
by hybrid distance geometry-dynamical simulated annealing calculations",
FEBS Lett.
229, 317-324.
- Nilges, M., Gronenborn, A.M., and Clore, G.M. (1989).
-
“Structure determination from
NMR conformational data by molecular dynamics calculations",
Proceedings of the
1989 Daresbury Study Weekend. Science and engineering research council,
Daresbury Laboratory, Warrington, WA4AA0, UK.
- Nilges, M., Habazettl, J., Brünger, A.T., and Holak, T.A. (1991).
-
“Relaxation Matrix Refinement of the Solution Structure of Squash
Trypsin-Inhibitor", J. Mol. Biol. 219, 499-510.
- Nilges, M., Kuszewski, J., and Brünger, A.T. (1991).
-
In: Computational Aspects of the Study of Biological
Macromolecules by NMR, (J.C. Hoch, ed.). New York: Plenum Press,
pp. 451-455.
- Nilsson, L., Clore, G.M., Gronenborn, A.M., Brünger, A.T., and Karplus, M. (1986).
- “Structure refinement of
oligonucleotides by molecular
dynamics with nuclear Overhauser effect interproton distance restraints:
Application to 5' d(C-G-T-A-C-G)
 ",
J. Mol. Biol. 188, 455-475.
",
J. Mol. Biol. 188, 455-475.
- Nilsson, L. and Karplus, M. (1986).
- “Empirical Energy
Functions for Energy Minimization and Dynamics of
Nucleic-Acids", J. Comp. Chem. 7, 591-616.
- Nordman, C.E. (1980).
- “Procedures for Detection and
Idealization of Non-crystallographic Symmetry with
Application to Phase Refinement of the Satellite
Tobacco Necrocis Virus Structure", Acta Cryst. A36, 747-754.
- Pearlman, D.A. and Kollman, P.A. (1991).
- “Are Time-Averaged
Restraints Necessary for Nuclear Magnetic Resonance Refinement? A
Model Study for DNA",
J. Mol. Biol. 220, 457-479.
- Powell, M.J.D. (1977).
- “Restart Procedures for the
Conjugate Gradient Method", Mathematical Programming
12, 241-254.
- Rao, S.N., Jih, J.-H., and Hartsuck, J.A. (1980).
-
“Rotation-Function Space Groups", Acta Cryst. A36, 878-884.
- Rieping, W., Habeck M., and Nilges, M. (2005).
- “Modeling errors in NOE data
with a lognormal distribution improves the quality of NMR structures”.
J. Am. Chem. Soc. 127, 16026-16027.
- Rogers, D. (1965).
- “Statistical Properties of Reciprocal Space",
In "Computing Methods in
Crystallography" (Rollett, J.S., ed.), 117-132.
- Rossmann, M.G. and Blow, D.M. (1962).
- “The Detection of
Sub-Units Within the Crystallographic Asymmetric Unit",
Acta Cryst. 15, 24-31.
- Ryckaert, J.-P., Ciccotti, G., and Berendsen, H.J.C. (1977).
-
“Numerical-Integration of Cartesian Equations of Motion of a
System with Constraints - Molecular-Dynamics of N-Alkanes",
J. Comput. Phys. 23, 327-341.
- Sass, H.-J., Musco, G., Stahl, S.J., Wingfield, P.T., and Grzesiek, S. (2001).
- “An easy way to include weak alignment constraints into NMR
structure calculations",
J. Biomol. NMR 21, 275-280.
- Schneider, T., Brünger, A.T., and Nilges, M. (1999).
- “Influence of
internal dynamics on accuracy of protein NMR structures: Derivation of
realistic model distance data from a long molecular dynamics
trajectory”, J. Mol. Biol. 285, 727-740.
- Schomaker, V., Waser, J., Marsh, R.E., and Bergman, G. (1959).
-
“To Fit a Plane or a Line to a Set of Points by
Least Squares", Acta Cryst.
12, 600-604.
- Schwieters, C.D. and Clore, G.M. (2001).
- “Internal Coordinates for
Molecular Dynamics and Minimization in Structure Determination and
Refinement",
J. Magn. Reson. 152, 288-302.
- Solomon, I. (1955).
- “Relaxation Processes in a System of 2
Spins", Phys. Rev. 99, 559-565.
- Stout, G.H. and Jensen, L.H. (1989).
- X-ray Structure
Determination, Wiley, New York.
- Strong, R., (1990).
- Ph.D. diss., Harvard University.
- Tarjan, R. (1983).
- Data Structures and Network Algorithms.
Society for Industrial and Applied Mathematics, Philadelphia.
- Tjandra, N., Garrett, D.S., Gronenborn, A.M., Bax, A., and Clore, G.M. (1997a).
- “Defining long range order in NMR structure determination
from the dependence of heteronuclear relaxation times on rotational
diffusion anisotropy",
Nature Struct. Biol. 4, 443-449.
- Tjandra, N., Omichinski, J.G., Gronenborn, A.M., Clore, G.M., and Bax, A. (1997b).
- “Use of dipolar 1H-15N and 1H-13C couplings in the
structure determination of magnetically oriented macromolecules in
solution",
Nature Struct. Biol.
4, 732-738.
- Tjandra, N., Marquardt, J., and Clore, G.M. (2000).
- “Direct
refinement against proton-proton dipolar couplings in NMR structure
determination of macromolecules",
J. Magn. Reson. 142, 393-396.
- Torda, A.E., Scheek, R.M., van Gunsteren, W.F. (1989) ;
- SPMldquo;Time-Dependent
Distances Restraints in Molecular Dynamics Simulations",
Chem. Phys. Letters 157, 289-294.
- Torda, A.E., Scheek, R.M., van Gunsteren, W.F. (1990) ;
- SPMldquo;Time-Averaged
Nuclear Overhauser Effect Distance Restraints Applied to Tendamistat",
J. Mol. Biol. 214, 223-235.
- Treutlein, H., Schulten, K., Deisenhofer, J., Michel, H., Brünger, A.T., and Karplus, M. (1992).
- “Chromophore-Protein Interactions
and the Function of the Photosynthetic Reaction Center:
A Molecular Dynamics Study",
Proc. Natl. Acad. Sci. USA 89, 75-79.
- Verlet, L. (1967).
- “Computer Experiments on Classical Fluids. I.
Thermodynamical Properties of Lennard-Jones Molecules",
Phys. Rev. 159, 98-105.
- Vuister, G., Delaglio, F., and Bax, A. (1993).
- “The use of
 J
J
 coupling constants as a probe
for protein backbone conformation"
J. Biomol. NMR
3, 67-80.
coupling constants as a probe
for protein backbone conformation"
J. Biomol. NMR
3, 67-80.
- Wang, H., and Stubbs, G. (1993).
- “Molecular dynamics in refinement
against fiber diffraction data",Acta Cryst A49, 504-513.
- Weiner, S., Kollman, P., Case, D., Singh, U.C., Ghio, C., Alagona, G., Profeta, S., and Weiner, P. (1984).
- “A new force field for molecular
mechanical simulation of nucleic acids and proteins",
J. Am. Chem. Soc. 106, 765-784.
- Weis, W. I., Brünger, A. T., Skehel, J. J., and Wiley, D. C. (1990).
-
“Refinement of the Influenza Virus Haemagglutinin by Simulated
Annealing", J. Mol. Biol. 212, 737-761.
- Welsh, L.C., Symmons M.F., and Marvin D.A., (2000).
- “The molecular structure
and structural transition of the
 -helical capsid in filamentous
bacteriophage Pf1", Acta Cryst. D56, 137-150.
-helical capsid in filamentous
bacteriophage Pf1", Acta Cryst. D56, 137-150.
- Wilson, A.J.C. (1949).
- “The Probability Distribution of X-ray Intensities",
Acta Cryst. 2, 318-321.
- Wolfram, S. (1991).
- Mathematica, A System for Doing Mathematics
by Computer, 2nd Ed. Redwood City, California:
Addison-Wesley Publishing Company, Inc.
- Wu, Z., Tjandra, N., and Bax, A. (2001).
- “
 P chemical shift
anisotropy as an aid in determining nucleic acid structure in liquid
crystals",
J. Am. Chem. Soc.
123, 3617-3618.
P chemical shift
anisotropy as an aid in determining nucleic acid structure in liquid
crystals",
J. Am. Chem. Soc.
123, 3617-3618.
- Yip, P. and Case, D. A. (1989).
- “A New Method for
Refinement of Macromolecular Structures based on Nuclear
Overhauser Effect Spectra", J. Magn. Reson. 83, 643-648.
Xplor-NIH 2025-11-07
